How to implement a model using cuDIMOT:
5 Steps:
1. Choose a model name and create model directory
2. Set the number and size of the model parameters
3. Define the model functions
4. Define the model priors
5. Compile
1. Choose a model name and create model directory
Set the enviroment variable modelname with the name of your model. This name will be used for generating binaries files at compilation time.
export modelname=NEWMODEL
Run to the script createModel.sh
./createModel.sh
This script will create a directory and several files needed by your model in mymodels/$modelname
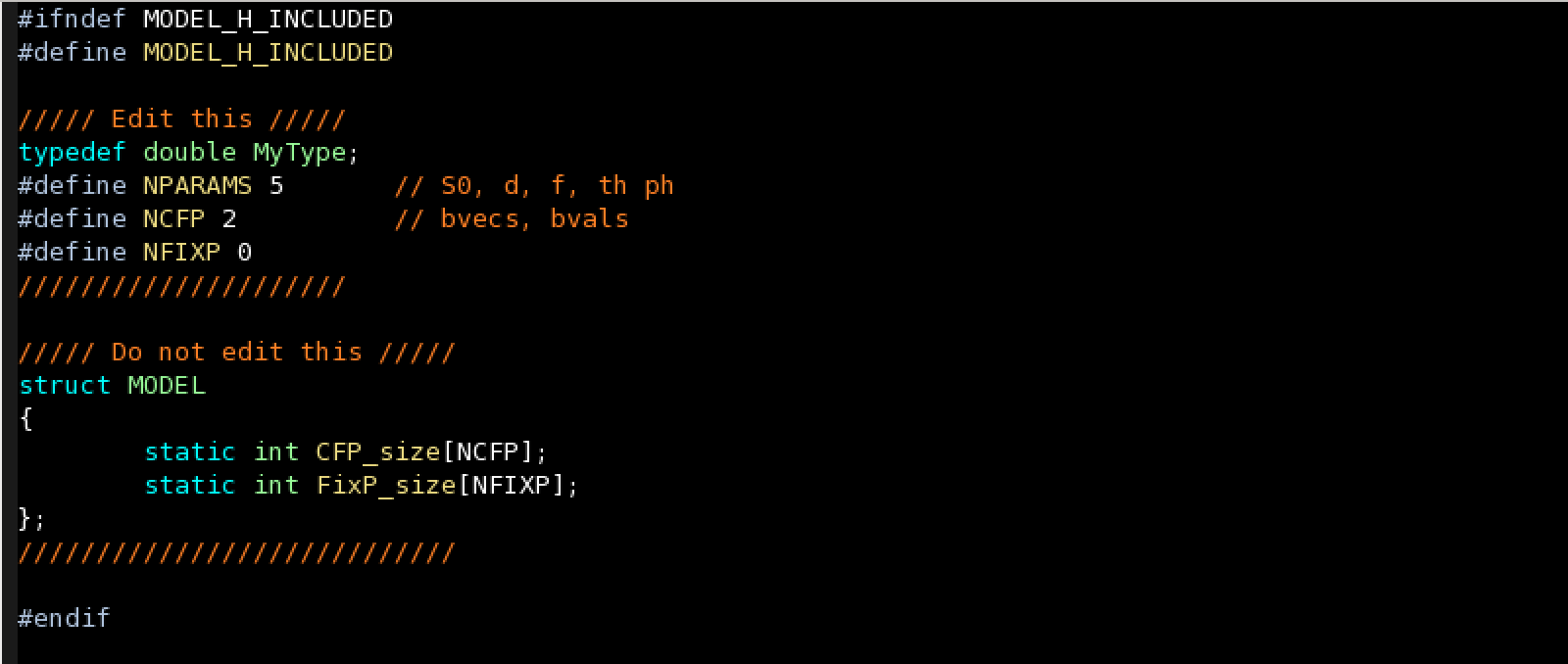
2. Set the number and size of the model parameters
You need to edit the file mymodels/$modelname/modelparameters.h for specifying the number of parameters.
You can choose between float or double types (single/double precision). The recommendation is to use double type for complex models.
There are 3 classes of parameters:
- Parameters to be estimated during the fitting process, defined with NPARAMS.
- Common Fixed Parameters (CFP): Parameters that are fixed during the fitting process and are common to all the voxels, defined with NCFP. In the above example there are 2 CFP, bvecs and bvals.
- Fixed Parameters (FixP): Parameters that are fixed during the fitting process and are different for each voxel (for instance, T1 or S0 in some models). The number of these parameters is defined with NFIXP.
For setting the size of the CFP and FixP you need to edit the file mymodels/$modelname/modelparameters.cc.
- CFP_size: You must specify the size of each CFP. The size of a Common Fixed Paramete is: [Data_Measurements x K]. Only K must be provided. In the above example there are 2 CFP, bvecs with size [Data_Measurements x 3] and bvals with size [Data_Measurements x 1].
- FixP_size: As in the previous case, a size must be provided for each Fixed Parameter, but in this case the size is [Number_Voxels x K] and only K (size of 4th dimension of the volume) must be provided.
3. Define the model functions
The file containing the model fuctions is mymodels/$modelname/modelfunctions.h
This file must contain 5 functions:
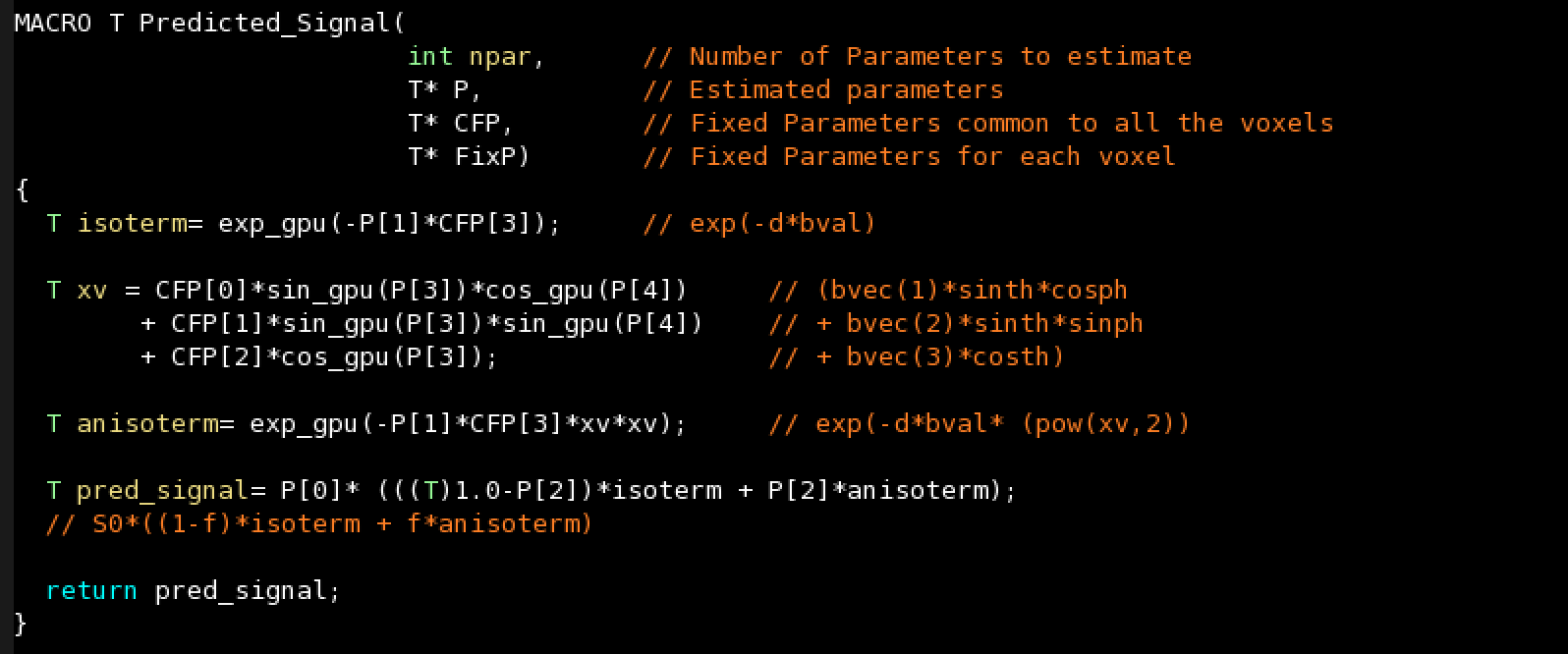
- The model predicted signal
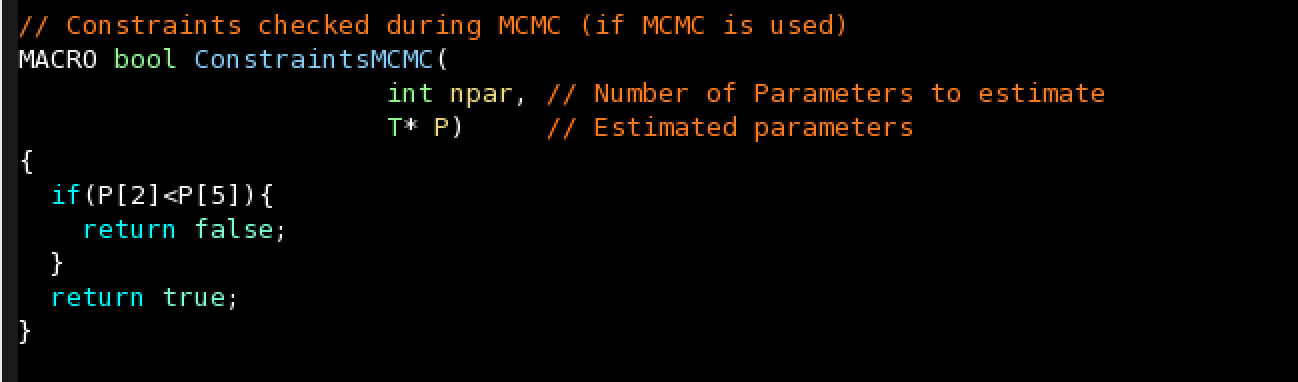
- Constraints during MCMC
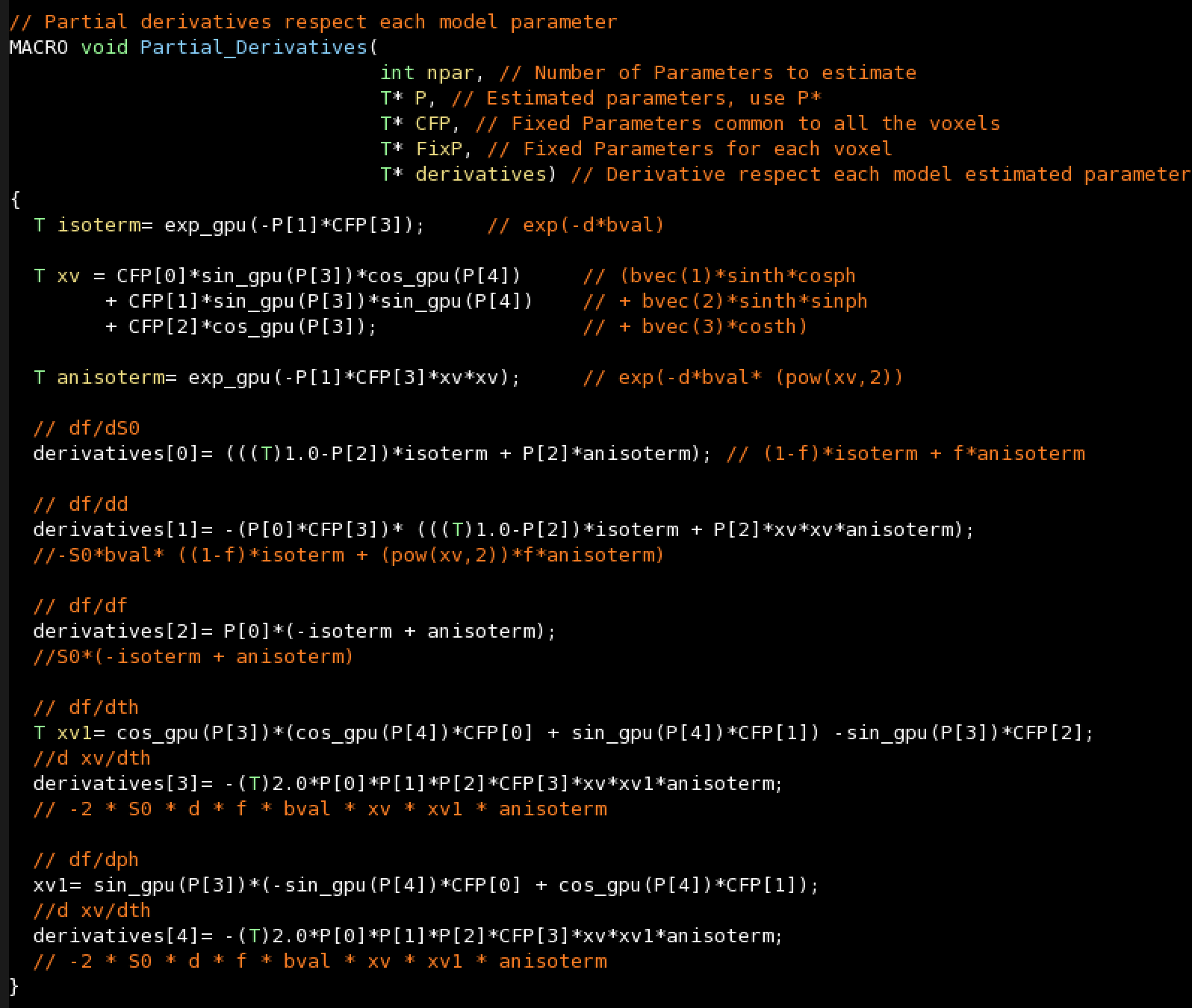
- Partial derivatives for Levenberg-Marquardt
- Constraints after Levenberg-Marquardt
- Custom Voxelwise Priors (for MCMC)
The functions should receive and return always the same arguments and use the undefined type T (you should not modify this).
The functions can call subroutines written in this file or defined in different files using #include. Subroutines must be defined before beeing called. All the functions and subroutines should be written as C-CUDA functions and use MACRO when they are defined.
- Do not change the name of the functions.
- Do not change the function parameters (although you can use you own parameter names in subroutines).
- If you use the suffix _gpu for the mathematical operations, the tool will select automatically the correct precision (single or double) depending on the datatype T.
- For obtaining a good performance, use the values stored in the arrays P, CP, CFP and derivatives as much as possible (for reading), but do not overwrite them. Avoid the declaration of new variables if is possible, particulary arrays.
The model predicted signal: Used during Grid-Search, Levenberg-Marquardt and MCMC. Return a single value of type T.
Constraints during MCMC: Return false if a constraint is not satisfied during MCMC and the sample will be rejected. By default, it should return true.
For instance, if you want to ensure that the fifth parameter is always lower than the third:
Partial derivatives for Levenberg-Marquardt: From the model predicted signal (Not from the cost function).
It returns an array of values, one for each parameter of the model. If you are not going to use Levenberg–Marquardt algorithm for fitting your model (--runLevMar=false), this function can be empty (but the function must be declared).
You can use numerical differentiation by using the function NUMERICAL(num_param). For instance:
derivatives[2]=NUMERICAL(2);
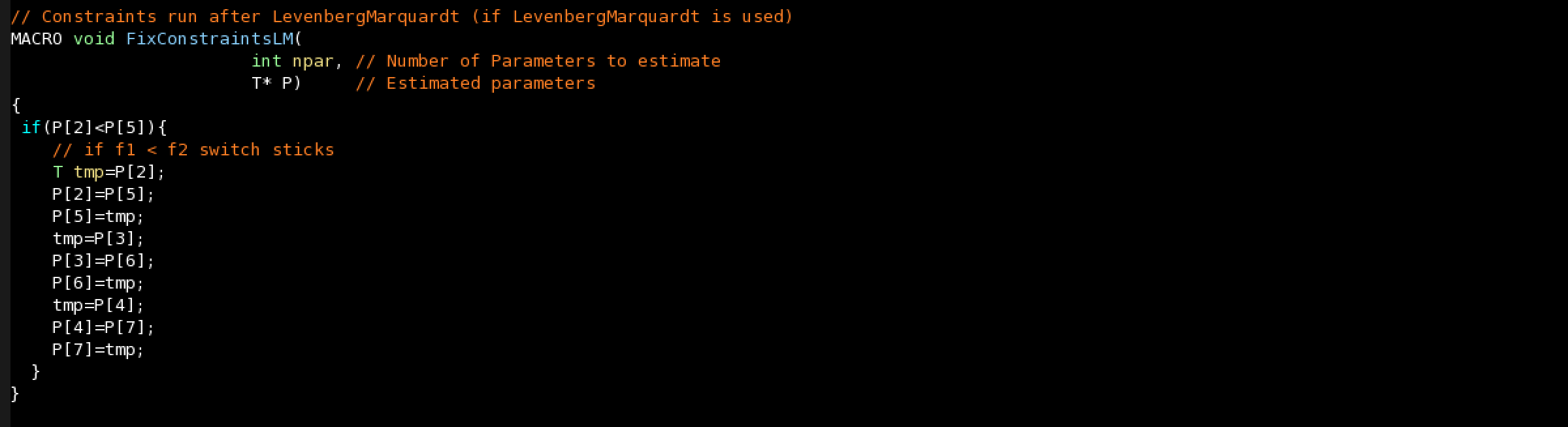
Constraints after Levenberg-Marquardt: You can specify what to do with some parameters after Levenberg-Marquardt if a constraint is not satisfied.
For instance, if your model has two sticks with their volume fractions in P[2] and P[5] and you want the first stick to have a higher fraction (P[3],P[4] are the angles parameters of the first stick and P[6],P[7] the angles of the second stick):
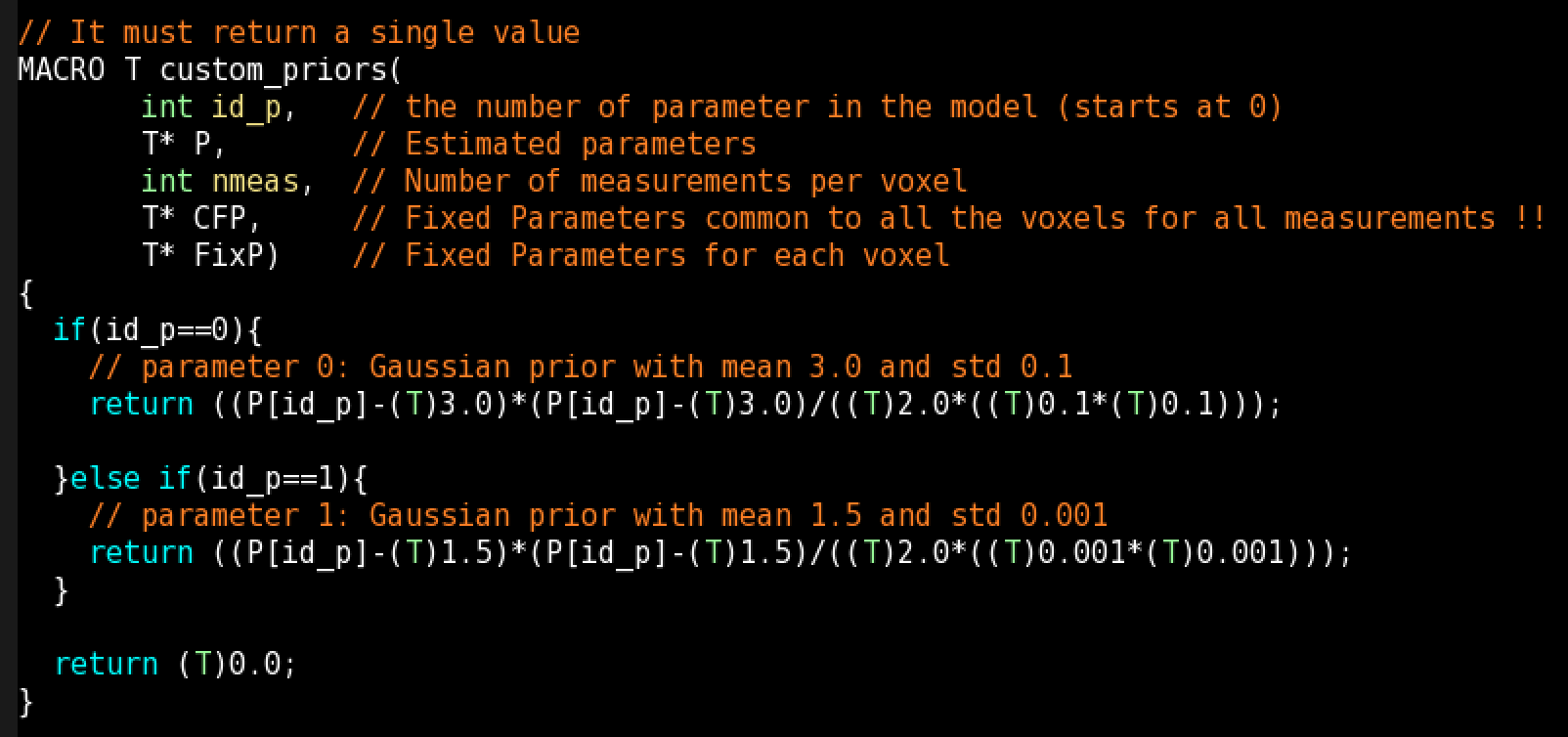
Custom Voxelwise Priors (for MCMC): With this function you can define your own priors for any parameter. If you define the prior as custom() (see next section), this function will be called. You can use voxelwise information using FixP (Fixed Parameters).
You can write a different custom prior for each model parameter, but you must use the id_p (Id of parameter) argument and if-else-if conditions to differentiate between actions to take, since any prior defined as custom() will call this function:
4. Define the model priors
Although this information is provided at execution time, you can define model priors in a file for later use. For example, you can use the file mymodels/$modelname/modelpriors.h.
All the priors are optional, but a file must be provided at execution time.
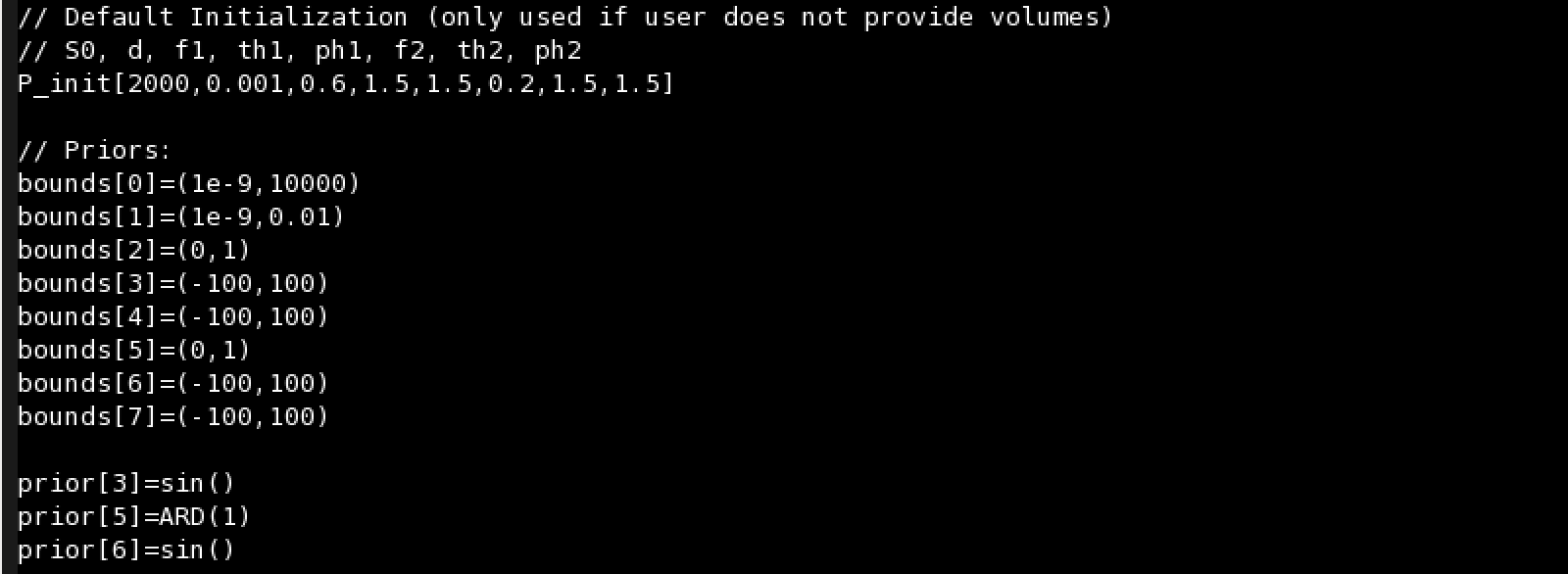
You can specify a default initilization value for each parameter. However, volumes with values for each voxel can be provided at execution time.
You can provide three kind of bounds for each parameter:
- Lower and Upper bounds: bounds[number_parameter]=(lower,upper)
- Only lower bound: bounds[number_parameter]=(lower,)
- Only upper bound: bounds[number_parameter]=(,upper)
The bounds will be taken into account during Grid-Search, Levenberg-Marquardt (using automatic transformations) and MCMC.
You can provide five kind of priors for each parameter:
- prior[number_parameter]=Gaussian(mean,variance)
- prior[number_parameter]=Gamma(alpha,beta)
- prior[number_parameter]=ARD(fudge_factor)
- prior[number_parameter]=sin()
- prior[number_parameter]=custom()
The priors will be taken into account only during MCMC.
5. Compile
CUDIMOT uses FSL library, thus you need to set some FSL enviroment variables:
export FSLDIR=[Path to the main installed versions of the binaries/headers/scripts/libraries etc.] export FSLDEVDIR=[Path to your own development versions of the software] . $FSLDIR/etc/fslconf/fsl.sh export FSLCONFDIR=$FSLDIR/config export FSLMACHTYPE=`$FSLDIR/etc/fslconf/fslmachtype.sh`
Set path to CUDA toolkit. For instance:
export CUDA=/opt/cuda-7.5
Compile:
make install
Binaries files with the name of your model will be generated in $FSLDEVDIR/bin.